Overview of techniques involved (e.g. 'omics')
| Site: | EUPATI Open Classroom |
| Course: | Role of Pharmacogenetics / Pharmacogenomics in the Development of Medicines |
| Book: | Overview of techniques involved (e.g. 'omics') |
| Printed by: | Guest user |
| Date: | Friday, 11 July 2025, 4:48 AM |
1. Biomarker identification
(This section is organised in the form of a book, please follow the blue arrows to navigate through the book or by following the navigation panel on the right side of the page.)
Most biomarkers used in medicines research today are so-called molecular or imaging biomarkers:
- Molecular biomarkers may include genes, proteins (such as hormones or enzymes) and metabolites. This type of biomarker may require a sample to be taken from the patient. This could for example be blood, saliva or urine. The sample is then analysed, normally in a laboratory or with the use of a test kit. There are many good tests available that can accurately measure molecular biomarkers.
- Imaging biomarkers can be seen in an image, such as an X-ray or Magnetic Resonance Imaging (MRI). They do not require a sample to be taken from the patient.
There are many established biomarkers which are known to have clinical use.
In addition, many new biomarkers are being found and used during the development of new medicines. These can also either be molecular (e.g. 'omics’: genomics, proteomics and metabolomics) or imaging biomarkers. ‘Omics’ are described further in the following sections.
2. Genomics
The common definition of a genome is a person’s entire set of DNA (which contains their genes).- Most of the genome is in the nucleus or core of each cell. The nucleus acts as the control centre for the cell. It contains the genetic material and controls cell growth and reproduction.
- A small amount of the genome is in the mitochondria. These are small structures in the cell that are responsible for energy production.
The European Medicines Agency (EMA) definition of a genomic biomarker includes RNA as well as DNA:
“A measurable DNA and/or RNA characteristic that is an indicator of normal biological processes, pathogenic processes, and/or response to therapeutic or other interventions”
In other words, a genomic biomarker is a feature of DNA and/or RNA that can be measured to indicate a normal or disease process in the body. This might include an individual’s response to a medicine or other interventions.
2.1. Introduction to DNA and genetics
The basic process in genetics are:- DNA is transcribed into ‘messenger RNA’ (mRNA). Transcription means that the information from a strand of DNA is essentially copied into a new molecule of mRNA.
- mRNA is then translated into a protein.The information in mRNA is used as a template for building the protein.
This is known as the ‘central dogma’ of molecular biology. It describes a process called gene expression.
DNA transcription and
translation
Figure 1: DNA makes RNA makes protein. Taken from http://www.atdbio.com/content/14/Transcription-Translation-and-Replication
DNA is made up of a double strand of molecules called ‘bases’:
- Each base is attached to another base on the other strand, making a ‘base pair’.
- The bases come in four types. These are given the letters A (adenine), T (thymine), G (guanine) and C (cytosine).
- It is the order of these bases that makes up the DNA sequence. The pairing of the bases links the two strands of DNA together into a double helix shape. A can only link with T, and G can only link with C.
DNA Helix and base
pairs
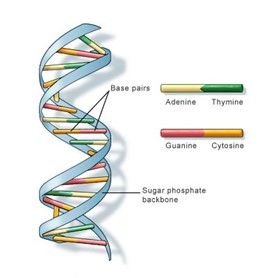
Figure 2: model of DNA, featuring
four chemical ‘letters‘- adenine, thymine, guanine and cytosine, or A, T, G and C. Taken from the US National Library of Medicine.
The sequence of the bases is important because this is the set of instructions the body uses to build proteins. Proteins are used in every cell. They ensure that each part of the body does what it needs to do to allow us to grow and function.
A gene is a length of DNA that contains the code for a particular protein. The structure of any given protein depends largely on the sequence of bases in a gene or genes. Any change to the DNA sequence, such as a genetic variant or mutation, may affect the production of a normal protein. This may in turn affect the normal function of a cell or tissue.
Mike Gilchrist from the UK’s Medical Research Council (MRC) writes:
“Can we get a feel for how much ‘information’ the genome (the entire set of DNA in an individual) contains? In simple terms a good thick airport novel contains about a million letters, and so a library of 3.000 such books would hold the same amount of ‘text’ as the human genome. This would fit easily into bookcases lining one wall of a generously sized living room. If we were to devote our leisure time to reading these ‘books’, and could get through one a week, it would take sixty years to plough through our entire genome..”.
“…our genome sequence is largely made up of repetitive, or uninformative, sequence [that is, not gene sequences], which we think of as having little impact on the day to day running of our bodies. This material is sometimes called ‘junk’ DNA… Our genes, which do most of the useful work, occupy the remaining ‘interesting’ 2% of the genome, and are the key to our biology. We have about 20.000 to 25.000 genes, and each is responsible for making one of the many smaller molecules (mostly proteins) that we need to grow and to function… In addition, the sequence around a gene contains signals which, in concert with other genes, tell the body where and when to produce that gene’s protein. This is the origin of our need as biologists to sequence the genome, as only this way can we begin to understand the functioning of all these genes.”
2.2. DNA Sequencing
DNA sequencing is a process used to find out the exact order of bases in a strand of DNA. For many years, a technique called capillary electrophoresis (CE) DNA sequencing (or Sanger sequencing) was used in laboratories all over the world.
The human genome has about 3 billion (3.000 million) base pairs. To sequence the first full human genome took over 10 years and the first draft was published in 2001.
Follow this link to see a video which illustrates the principle of the Sanger method of DNA sequencing:
Source: http://www.dnalc.org/view/15479-Sanger-method-of-DNA-sequencing-3D-animation-with-narration.html
A new method has been found since the first human genome was sequenced. This is called next generation sequencing (NGS). This process is a lot faster and has allowed much more rapid research progress. The basic chemistry is the same as for Sanger sequencing. However the technique allows millions of tiny fragments of DNA to be sequenced at the same time, rather than just a few hundred.
2.3. DNA as a Biomarker
Why would we want to sequence other humans’ genomes if we have already sequenced one? The answer is that much of what makes humans ‘individuals’ is determined by many small differences in our DNA. Small differences in our DNA often still result in working proteins. Yet, some variations stop a protein being produced or lead to the production of a protein that does not work properly. Some of these variations are explained later in the text.
Mutations are variations in DNA sequence. These can occur, for example, when sections of DNA are lost (deleted), or inserted into new positions within a length of DNA. Mutations can affect the structure of the protein that a gene encodes. In some cases, they can therefore affect the health of an individual. Mutations are relatively rare - they are only found in a few people.
For example, cystic fibrosis is caused by mutations in the CFTR gene. As explained in Figure 3, this affects the function of a protein which helps produce mucous in the lungs and in other parts of the body. The mucous becomes unusually thick and sticky, and this leads to breathing problems and bacterial infections in the lungs.
CFTR protein channel
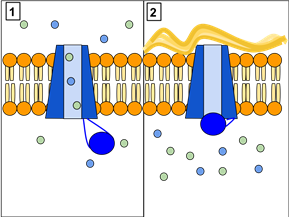
Figure 3: The CFTR protein is a channel protein that controls the flow of H2O and Cl- ions in and out of cells inside the lungs. Panel 1 shows a CFTR protein which works correctly. As you can see, ions flow freely in and out of the cells. Panel 2 shows a CFTR protein that does not work as it should. Here, theions cannot flow out of the cell because a channel is blocked. This causes cystic fibrosis, which leads to a build-up of thick mucus in the lungs. Created by Lbudd14, under Creative Commons CC BY-SA 3.0 (http://en.wikipedia.org/wiki/Cystic_fibrosis#mediaviewer/File:CFTR_Protein_Panels)
SNPs (single nucleotide polymorphism) are the simplest form of variation in DNA sequence. This is a change in a single base pair, for example a length of DNA might read AATGC and a SNP might change this to AGTGC. SNPs are relatively common and they are found in many people. They can occur within genes and in other parts of the DNA sequence that do not contain information to build up proteins. Most SNPs do not affect an individual’s health.
SNP DNA molecule
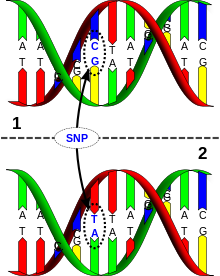
Figure 4: DNA molecule 1 differs from DNA molecule 2 at a single base-pair location (in this case, it is a C/T polymorphism). Created by David Hall, under Creative Commons licence CC BY 2.5 (http://en.wikipedia.org/wiki/Human_genetic_variation#mediaviewer/File:Dna-SNP.)
Researchers can perform genome wide association (GWA) studies by using whole genome sequencing. GWA studies link variations between people with differences in their health, disease and their response to treatment.
GWA studies can identify DNA variants that are useful as biomarkers. Other studies that look at specific ‘candidate’ genes can also do this. These are studies that look at the link between a specific gene and disease. They are more focused than a GWA study that looks at the whole genome. This is becoming a very fruitful area of research, even though the relationship between specific variants and disease is not simple in most cases. Often, there are many variants associated with a disease, not just one. In addition, the impact of epigenetic changes can complicate the picture. Epigenetic changes are changes in the way that genes are switched on and off without a change in the DNA sequence.
2.4. Gene Expression as a Biomarker
DNA sequencing will tell you about the genetic variants that individual persons have. But it is also important to know which genes are being ‘expressed’, or switched on, and how strongly. When genes are expressed, they are first transcribed into mRNA. They are then translated into proteins. See introduction to this section 'Overview of techniques involved'.
mRNA molecules are made up of a single strand of bases (unlike DNA which is a double strand). Levels of mRNA can be measured to give a gene expression signature. The whole set of mRNA molecules in a cell at any one time is sometimes referred to as the transcriptome. Transcriptomics is used to learn more about how genes are turned on in different types of cells.
Gene expression signatures can vary over time and between different tissues and different parts of the body.
For example, a recent study into advanced ovarian cancer has shown that it is possible to separate patients into those who respond well to platinum chemotherapy and those who do not. This is shown by looking at how genes are expressed differently in their tumours.
Platinum chemotherapy has several common serious side effects. If a doctor knows how likely a patient is to respond, it helps the doctor and the patient assess if it is worth risking these side effects.
How gene expression is measured
Microarray technology is used to identify gene expression signatures. mRNA molecules are collected and converted into artificial molecules called ‘complementary DNA’ (cDNA). cDNA is much more stable and easier to work with than mRNA. The cDNA molecules are then put on a glass slide that has thousands of DNA sequences already bound to it. The cDNA will stick if a cDNA sequence matches a bound piece of DNA.
The result is a microarray. This is a pattern of fluorescent (lit up) dots of different colours on the slide. The colours show how strongly different genes in the sample are expressed compared to another sample which is used as a ‘control’. For example, you may compare a cancer cell sample to a normal cell from the same person which would then work as a control. One colour means high levels of expression, while a second colour means low levels. Another will indicate the same level of expression between the sample and control.
Computers are used to measure and analyse the fluorescence pattern.
Gene expression profile

Figure 5: Gene expression profile measured with microarray technology. Taken from http://www.biomedcentral.com/content/pdf/1752-0509-5-161.pdf
NGS technology (described in section 2.2) can also be used to measure mRNA. It is therefore also used in gene expression studies. Compared to microarrays, it is able to detect more subtle changes in gene expression, which might be important in disease processes.
2.5. Epigenetics
Epigenetics is the study of how genes can be turned on or off, or modulated (e.g. turned up or down), without any changes to their DNA sequence (i.e. without a mutation). The study of epigenetics is growing and is important in our understanding of the risk of getting a disease, how a disease will progress, or how it will respond to treatment.
Epigenetic changes are made by chemical changes in the DNA. They can occur in response to changes within the body, or to external factors such as UV light, diet, tobacco use or stress.
Epigenetic changes also happen when there are changes to the protein molecules that bind to DNA. Many of these proteins are involved in making epigenetic changes, and in ‘reading’ the resulting DNA. The pharmaceutical industry is getting more and more interested in such proteins, because they may be suitable new targets for medicines. Indeed, several medicines that influence epigenetic changes are already in clinical development and some have a marketing authorisation (such as ‘azacytidine’ for myelodysplastic syndromes)
Many cancers are related to epigenetic changes for specific genes. The epigenetic regulation of gene expression is called ‘gene silencing’. Silencing means that a cell can prevent a certain gene to be expressed. Methods to silence genes are increasingly used to produce therapeutics that can combat cancer and diseases. An example is the epigenetic silencing of the ‘GSTP1’ gene (the gene’s expression is reduced). This gene is likely to be a cause of prostate cancer.
Most progress to date in the discovery of medicines within epigenetics has been for cancer treatments. For example for breast and liver cancers, leukaemias and Hodgkin’s lymphoma.
3. Proteomics
Proteomics provides information about the proteome. A proteome is the entire set of proteins in an organism, cell or tissue. Proteomics also provides information on protein levels, any variations and changes they have, and which molecules they interact with.
The process of protein production using mRNA molecules is called translation, which consists in various steps mentioned previously in this topic:
- mRNA molecules are produced from the DNA in genes.
- The mRNA molecules in turn are used to produce proteins.
- Therefore, the sequence of every protein depends on the sequence of a gene or genes.
Proteins are chains of amino acids, which fold into specific three-dimensional (3D) shapes. The sequence or order of the amino acids is determined by genetic sequence. However the protein’s final shape, and therefore what it does, can also be modified by chemical changes. This is known as post-translational modifications.
3D structure of Myoglobin
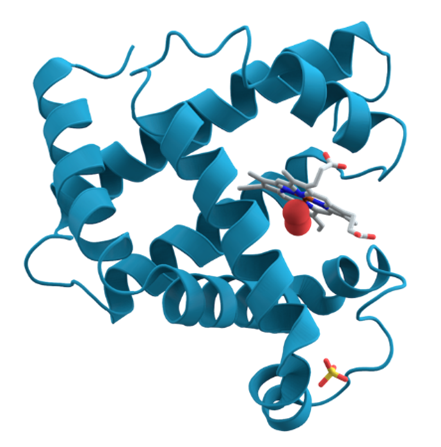
Figure 6: A 3D structure of the protein myoglobin (an oxygen-binding protein in the muscle) showing turquoise alpha helices. Towards the right-center among the coils, a group called a heme group (shown in gray) with a bound oxygen molecule (red). Created by AzaToth based on PDB entry, public domain http://en.wikipedia.org/wiki/Protein#mediaviewer/File:Myoglobin
3.1. The proteome
The proteome is the ‘end product’ of the genome. The genome hardly changes, but the proteome changes all the time in response to tens of thousands of signals from inside and outside the cell. For example, the proteome varies with:
- Health or disease.
- The nature of each tissue.
- The stage of cell development.
- Response to treatments.
The human body may contain more than 2 million different proteins, which each have different functions. Proteins serve vital functions in the body. They:
- Regulate cell division.
- Defend the body against disease - antibodies.
- Speed up metabolic reactions - enzymes.
- Transmit signals between brain cells - neurotransmitters.
As such, the proteome is often defined as the entire protein content in a given cell, tissue or organism at a certain point in time
3.2. Proteomics
The patterns of proteins can vary and researchers use these to find biomarkers that can help find new medicines. The patterns can be tied to a risk of getting a disease, how a disease will progress, or how it will respond to treatment.
New tests are underway and these tests can analyse thousands of protein levels and also look at their post-translational modifications (changes occurring after they have been assembled). The tests include technologies that are adjusted to be able to analyse proteins based on the molecule components. This allows for much faster analysis of proteomes.
4. Metabolomics
The metabolome is the entire set of small molecule metabolites in an organism, tissue or cell at any given time. The metabolome responds to many factors. These might include recent food intake, the menstrual cycle or the time of day. The metabolome
can change over a period of just seconds or minutes. Metabolomics helps us identify what effect a medicine is having. It is a new emerging field of ‘omics’ research and its purpose is to find and describe the small molecule metabolites in an organism:
- Small molecule metabolites are the molecules that make up the biochemistry of every cell.
- They are the intermediates and the products of metabolism.
- Their levels vary over time and they respond to thousands of internal and external factors.
Small molecule metabolites are below a certain size, which means they are able to cross cell membranes. Large molecules such as DNA, proteins and carbohydrates (such as starch) are therefore not included in this definition.
Metabolomics is now being used in some stages of medicines development even though it is a developing area. Metabolomic biomarkers can improve the non-clinical assessment of new medicines. They can also help in translational medicine, where exploratory research is translated into clinical research.
5. Imaging techniques
Imaging is another
group of techniques that can be used as biomarkers in medicines development.
Common imaging techniques in medicine are:
- Magnetic Resonance Imaging (MRI)
- Computed Tomography (CT)
- Ultrasound
- Positron Emission Tomography (PET)
The techniques are usually not invasive which means that no biological samples are collected. They can provide information about whole tissues, tumours or organs.
MRI and CT
measurements are often used in cancer patients to assess tumour size in Phase
III clinical trials. In 2011, the EMA gave a regulatory approval on the use of
MRI to select Alzheimer’s patients with early stage disease into clinical
trials . This was the first imaging biomarker for Alzheimer’s to be given a
positive opinion by a regulatory agency.
Very few imaging biomarkers have been qualified (i.e. validated) for early phases of development, although some non-validated methods can be used in exploratory clinical trials.
However, as with the newer ‘omics’approaches, much effort is put into research on imaging biomarkers for medicines development. For example, the Innovative Medicines Initiative (IMI) has helped to setup an international collaboration of academics and industry called QuIC-ConCePT (Quantitative Imaging in Oncology: Connecting Cellular Processes to Therapy, http://www.quic-concept.eu/). QuIC-ConCePT aims to develop imaging biomarkers that can show earlier and more accurately how patients’ tumours react in cancer clinical trials.
6. Bioinformatics
Bioinformatics helps scientists handle very large amounts of data and it is becoming an important area in medicine research. The UK’s Public Health Genetics (PHG) Foundation defines bioinformatics as a field that ‘combines concepts and knowledge from computer science, statistics and biosciences in order to manage, search, visualise and analyse biological and medical data’.
Modern ‘omics’ technologies can create very large amounts of complex data from single experiments. Thanks to this, many potential biomarkers can be assessed for a link with, for example, a particular tumour type or a disease sub-group. Scientists often study very large sets of samples (e.g. blood samples from patients and healthy donors) which are organised and stored in biobanks.
Without reliable analysis, ‘omics’ data would be of no use. Bioinformatics for ‘omics’ research is therefore an important and developing field. For example, pharmaceutical companies can search through massive amounts of genomic data to find links between specific genetic variants and diseases. Computer modelling uses available data to predict or ‘model’ certain situations. This method is used to help predict both the likely benefits and possible side effects of as development candidate before it is tested in patients.
7. Advanced therapy medicinal products
Conventional medicine depend on molecules (pharmaceuticals) to treat diseases and injuries. New therapies, called advanced therapy medicinal products (ATMPs) or simply ‘advanced therapies’, are promising. They are made from genes, cells or tissues and reseachers develop them for the treatment of a range of diseases and injuries.
The therapies aim to address the ‘root cause’ of diseases or injuries. In other words, they try to overcome problems with faulty genes, cells or tissues and they may have great potential for patients. As they are highly targeted therapies, they should help to move toward personalised medicine.
There are four main groups of advanced therapies:
- Gene therapy medicines
- Somatic-cell therapy medicines
- Tissue engineered medicines
- Combined advanced-therapy medicinal products.
The four groups will be explained below on the following subchapters.
7.1. Gene-therapy medicines
These medicines contain genes that have a therapeutic effect - they act to treat the disease. They work by inserting ‘recombinant’ genes into cells. Sometimes they use a virus (called a ‘vector’) to deliver the genes.
- A recombinant gene is a stretch of DNA that is created in the laboratory to have a certain sequence.
- When the gene is delivered into a cell, it causes the cell to either produce, or stop the production of a specific protein. By doing this, it can slow down or cure a disease.
- Diseases that have so far been targeted by gene therapy include genetic disorders, cancer and chronic (long-term) conditions.
Gene therapy using adenovirus vector
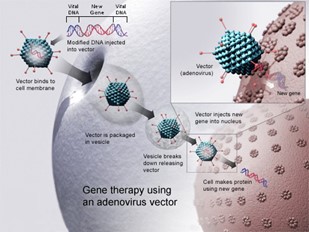
Figure 7: Gene therapy using an adenovirus vector. A new gene is inserted into a cell using an adenovirus. If the treatment is successful, the new gene will make a functional protein to treat a disease. Image created by the National Institutes of Health (accessed on https://en.wikipedia.org/wiki/Gene_therapy)
8. Somatic-cell therapy medicines
These medicines contain cells or tissues that have been biologically changed. Note that the cells are non-sex (non-germline) cells. The cells or tissues can be autologous (taken from the individual patient) or allogeneic (taken from another person, who might be related to the patient).
An example of somatic cell therapy is the use of a patient’s own immune cells. The cells are changed so that they can recognise and attack tumour cells when they are put back into the patient.
‘Stem cell’ therapy is one type of somatic cell therapy that has received widespread media attention. The goal of stem cell therapy is to replace tissues that are damaged due to illness or injury. This is done by ‘growing’ new tissue from stem cells (‘regeneration’). For example, the dead heart muscle found in heart failure, or the dopamine-producing cells damaged in Parkinson’s disease.
Another important use of stem cell therapy is haematopoietic stem cell transplantation (HSCT). The stem cells transplanted in this procedure are not changed, therefore HSCT is not referred to as somatic cell therapy.
- Haematopoietic stem cells in the bone marrow are essential for the production of blood cells.
- Chemotherapy and/or radiotherapy will partly or totally destroy the bone marrow of patients who are treated for blood or bone marrow cancers.
- HSCT is used to remove and then re-introduce haematopoietic stem cells into patients after such treatment, and allow them to start producing blood cells again.
There has been a dramatic increase in our knowledge about stem cells in the past few years. Also an increase in research and investment into their use as medicines. No medicines based on stem cells have yet received a marketing authorisation in the European Union (EU), but the EMA has offered advice to companies that develop these medicines for a number of years. Read more here.
Despite their promise, however, there is doubt about the possible drawbacks of using stem cells in medicines. Risks could be serious and unpreditable such as the development of tumours and rejection by the body.
The EMA has expressed concerns about patients who are seriously ill and who travel to clinics around the world for stem-cell-based treatments that are not authorised. Some clinics offer therapies at a high cost, without ethical approval, and there are serious concerns about their safety and efficacy. Read more here.
9. Tissue engineered medicines
These products contain cells or tissues that have been engineered (modified) so they can be used to repair, regenerate or replace tissue. Most tissue engineered products are cell-based, i.e. they use somatic cell therapy to build replacement tissue.
An example of a tissue engineered product is artificial skin which is used to treat patients with severe burns. Research into tissue engineered medicines is currently looking at cartilage, bone, nerve tissue, corneas and heart muscle.
Sometimes the term ‘tissue engineered medicine’ is used to mean a combined medicine.
10. Combined advanced-therapy medicinal products (combined ATMPs)
These medicines contain an advanced therapy plus one or more medical devices – for example, cells that are fixed in a matrix or structure to engineer or reconstruct tissue.
The EMA defines combined ATMPs as: “Combined ATMPs incorporate a cellular part consisting of cells or tissues and one or more medical devices or one or more active implantable medical devices as an integral part of the product”.
An example of a combined ATMP is the ‘autologous chondrocyte implantation’ (ACI) to repair joint cartilage (Figure 8). Patient cells (autologous chondrocytes) are grown in the laboratory until the required number of cells is obtained. The cells are planted onto a collagen membrane which is implanted into the damaged cartilage. The cells repair the damaged tissue, while the medical device (the collagen membrane) keeps the cells in place.
Preparation of
bioengineered chondrocyte cell sheets
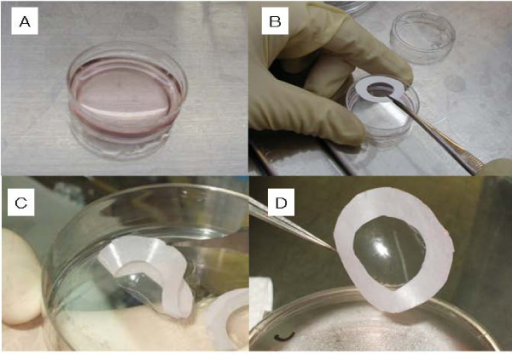
Figure 8: Preparation of bioengineered chondrocyte cell sheets for cartilage repair. A: dish containing growing cells. B: a membrane is placed on the sheet of cells and used to lift the cells out of the dish (C and D). Image courtesy of Mitani G et al., created under Creative Commons licence CC BY 2.0 http://openi.nlm.nih.gov/detailedresult.php?img=2662823_1472-6750-9-17-1&req=4